# Import all required libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats as stats
from scipy.stats import skewnorm
from scipy.stats import kurtosis, norm
from scipy.stats import gamma
import missingno as msno
import random
import statsmodels.api as sm
# Load in UK Smoking Data
births14 = pd.read_csv("data/births14.csv")
# Set seed
random.seed(123)Exploratory data analysis
Lecture 4
University of Arizona
INFO 511
Setup
Exploratory Data Analysis
What is exploratory data analysis?
Exploratory Data Analysis is a statistical approach to analyzing datasets to summarize their main characteristics, often using visual methods.
Examining data
| fage | mage | mature | weeks | premie | visits | gained | weight | lowbirthweight | sex | habit | marital | whitemom | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 34.0 | 34 | younger mom | 37 | full term | 14.0 | 28.0 | 6.96 | not low | male | nonsmoker | married | white |
| 1 | 36.0 | 31 | younger mom | 41 | full term | 12.0 | 41.0 | 8.86 | not low | female | nonsmoker | married | white |
| 2 | 37.0 | 36 | mature mom | 37 | full term | 10.0 | 28.0 | 7.51 | not low | female | nonsmoker | married | not white |
| 3 | NaN | 16 | younger mom | 38 | full term | NaN | 29.0 | 6.19 | not low | male | nonsmoker | not married | white |
| 4 | 32.0 | 31 | younger mom | 36 | premie | 12.0 | 48.0 | 6.75 | not low | female | nonsmoker | married | white |
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1000 entries, 0 to 999
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fage 886 non-null float64
1 mage 1000 non-null int64
2 mature 1000 non-null object
3 weeks 1000 non-null int64
4 premie 1000 non-null object
5 visits 944 non-null float64
6 gained 958 non-null float64
7 weight 1000 non-null float64
8 lowbirthweight 1000 non-null object
9 sex 1000 non-null object
10 habit 981 non-null object
11 marital 1000 non-null object
12 whitemom 1000 non-null object
dtypes: float64(4), int64(2), object(7)
memory usage: 101.7+ KB| fage | mage | weeks | visits | gained | weight | |
|---|---|---|---|---|---|---|
| count | 886.000000 | 1000.000000 | 1000.000000 | 944.000000 | 958.000000 | 1000.000000 |
| mean | 31.133183 | 28.449000 | 38.666000 | 11.351695 | 30.425887 | 7.198160 |
| std | 7.058135 | 5.759737 | 2.564961 | 4.108192 | 15.242527 | 1.306775 |
| min | 15.000000 | 14.000000 | 21.000000 | 0.000000 | 0.000000 | 0.750000 |
| 25% | 26.000000 | 24.000000 | 38.000000 | 9.000000 | 20.000000 | 6.545000 |
| 50% | 31.000000 | 28.000000 | 39.000000 | 12.000000 | 30.000000 | 7.310000 |
| 75% | 35.000000 | 33.000000 | 40.000000 | 14.000000 | 38.000000 | 8.000000 |
| max | 85.000000 | 47.000000 | 46.000000 | 30.000000 | 98.000000 | 10.620000 |
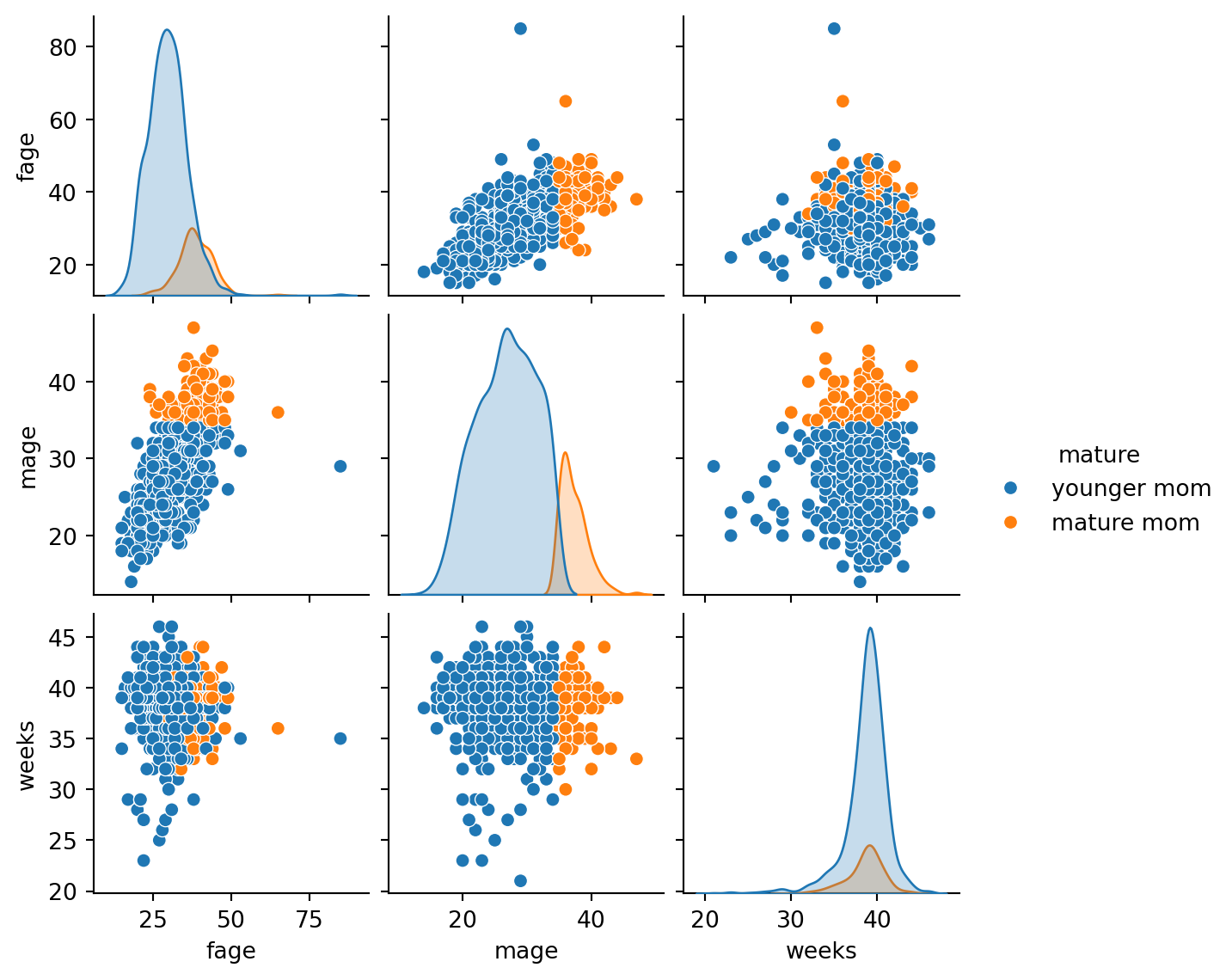
Visualizing data relationships
Group descriptive statistics
| fage | mage | ... | gained | weight | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | count | mean | ... | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | |
| premie | |||||||||||||||||||||
| full term | 775.0 | 30.967742 | 6.681591 | 15.0 | 26.0 | 31.0 | 35.0 | 49.0 | 876.0 | 28.329909 | ... | 38.0 | 98.0 | 876.0 | 7.434178 | 1.021699 | 3.93 | 6.77 | 7.44 | 8.0825 | 10.62 |
| premie | 111.0 | 32.288288 | 9.226826 | 15.0 | 27.0 | 32.0 | 36.0 | 85.0 | 124.0 | 29.290323 | ... | 41.0 | 85.0 | 124.0 | 5.530806 | 1.801182 | 0.75 | 4.50 | 5.75 | 6.5725 | 9.25 |
2 rows × 48 columns
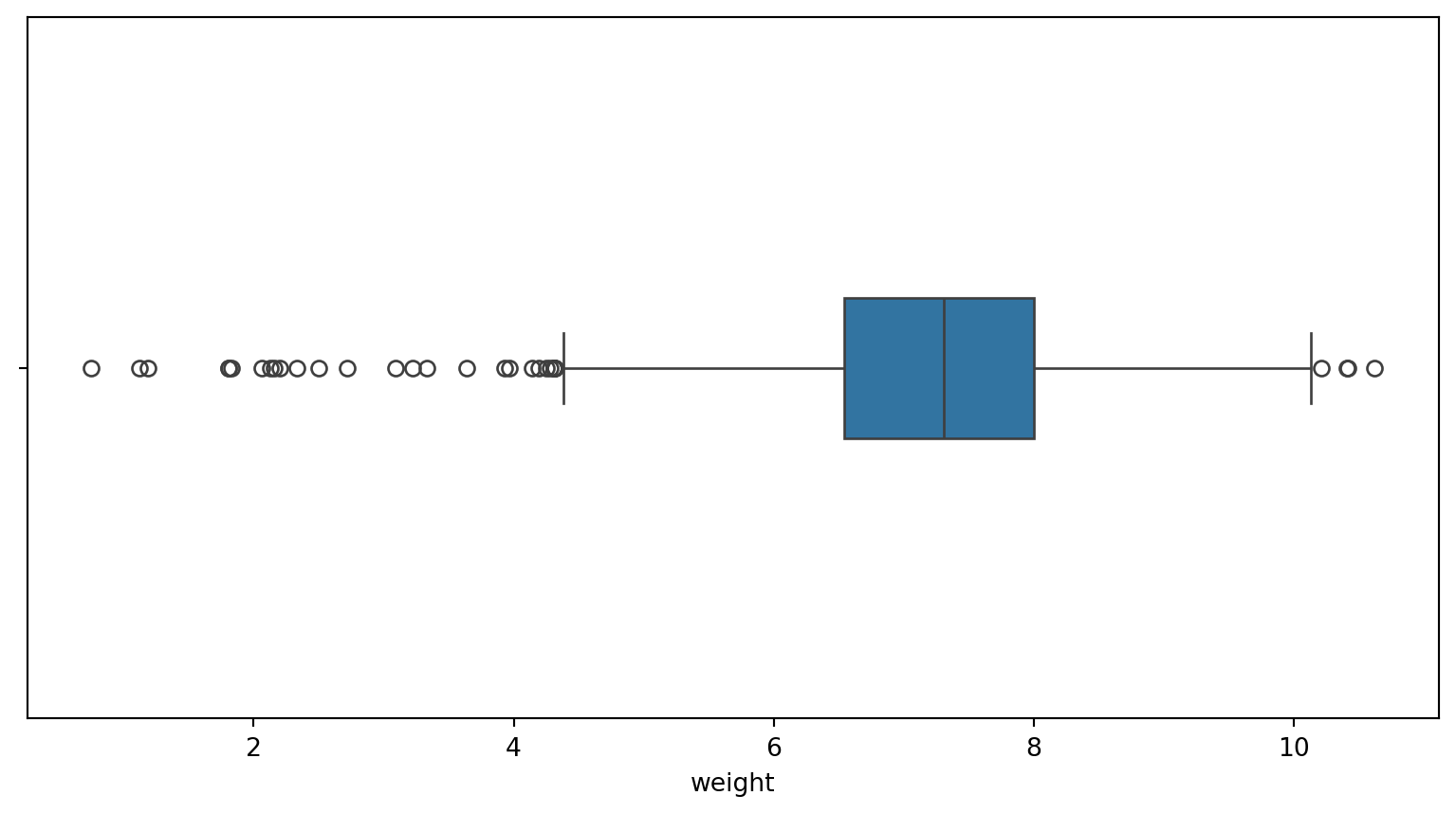
Outliers
Outliers are data points that are significantly different from others. Identifying and handling outliers is important in data analysis.
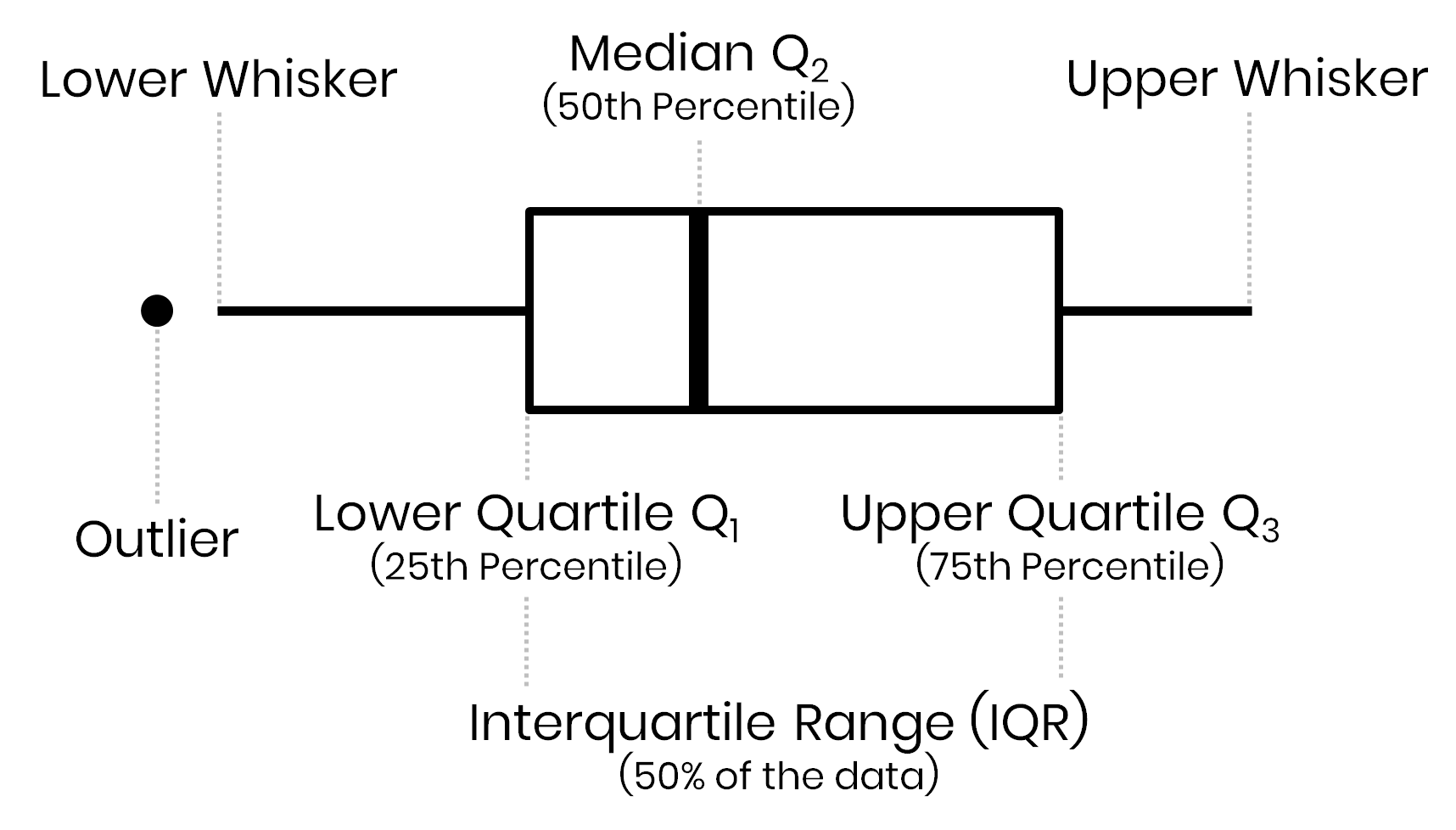
Outliers = 1.5 * Interquartile range
Assess outliers visually
Find outliers
fage: 7 outliers
mage: 1 outliers
weeks: 72 outliers
visits: 30 outliers
gained: 26 outliers
weight: 32 outliersfor column in births14.select_dtypes(include=np.number).columns:
q25 = births14[column].quantile(0.25)
q75 = births14[column].quantile(0.75)
iqr = q75 - q25
lower_bound = q25 - 1.5 * iqr
upper_bound = q75 + 1.5 * iqr
outliers = births14[(births14[column] < lower_bound) | (births14[column] > upper_bound)]
print(f"{column}: {outliers.shape[0]} outliers")q25: 1/4 quartile, 25th percentile;q75: 3/4 quartile, 75th percentileIQR: interquartile range, \(IQR = q75-q25\)lower;upper: lower, upper limit of \(1.5\times IQR\) used to calculate outliers
Remove outliers
# Select numerical columns
numerical_cols = births14.select_dtypes(include = ['number']).columns
for col in numerical_cols:
# Find Q1, Q3, and interquartile range (IQR) for each column
Q1 = births14[col].quantile(0.25)
Q3 = births14[col].quantile(0.75)
IQR = Q3 - Q1
# Upper and lower bounds for each column
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
# Filter out the outliers from the DataFrame
births14_clean = births14[(births14[col] >= lower_bound) & (births14[col] <= upper_bound)]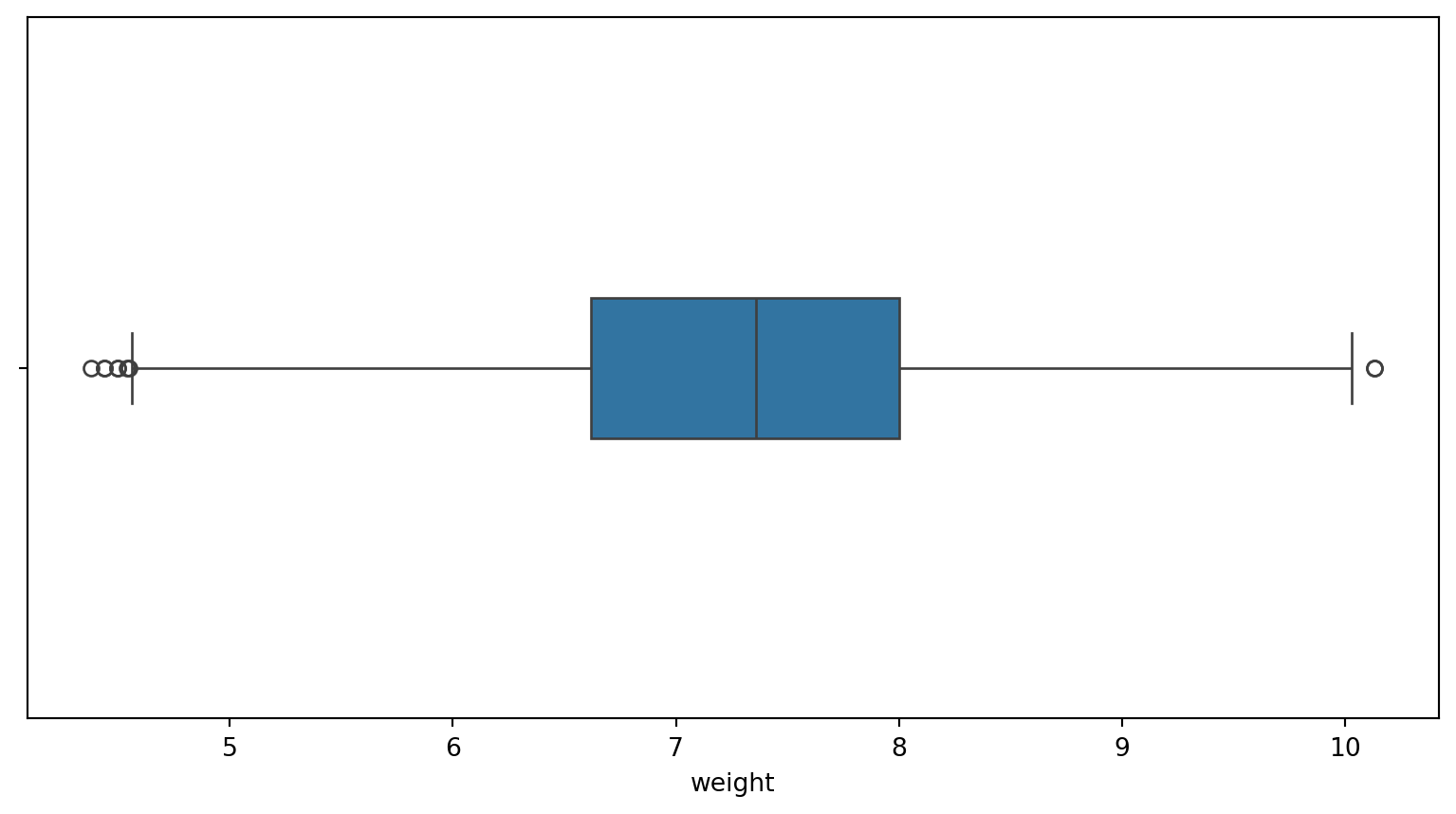
Why are there still outliers?
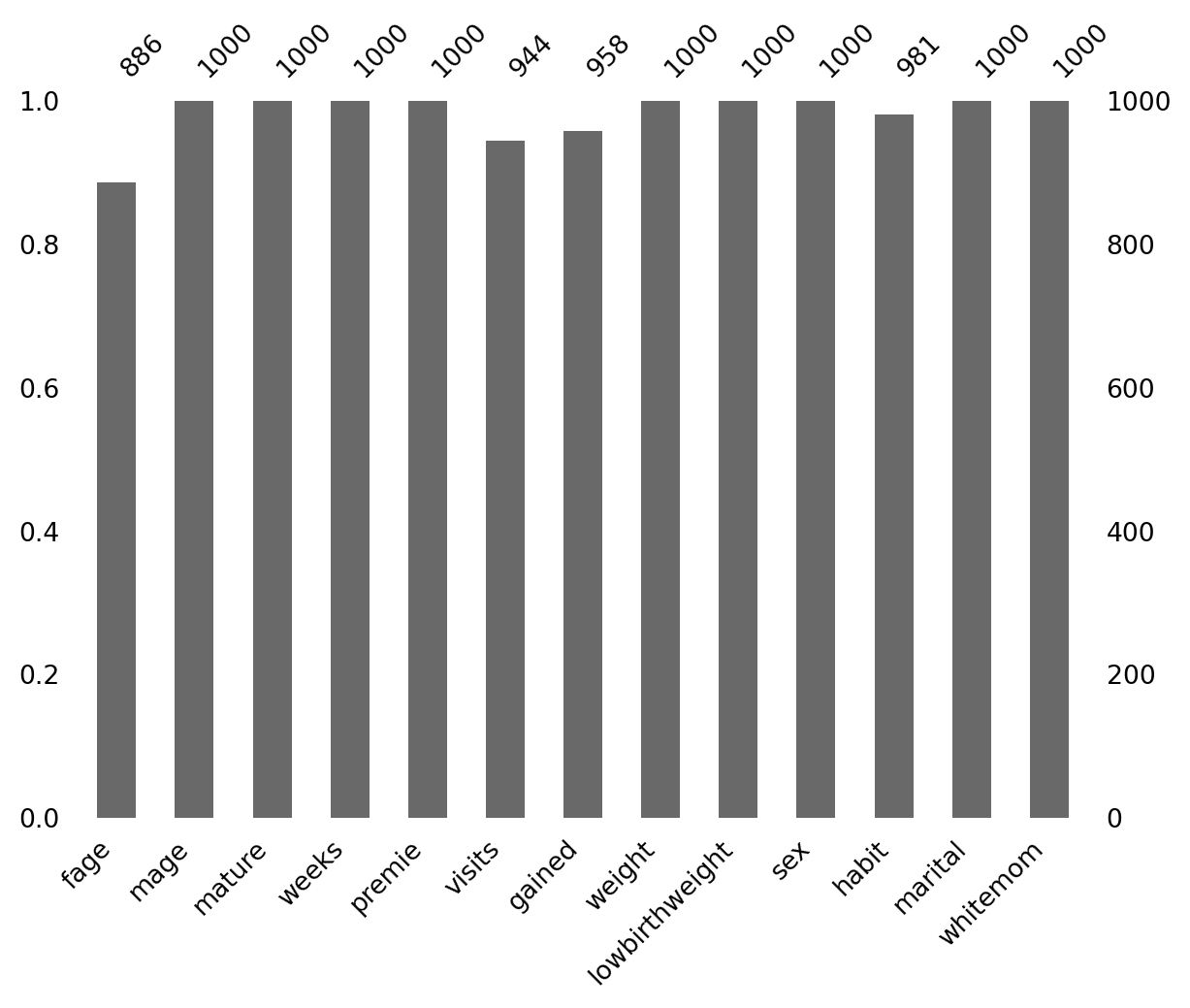
Missing values (NaN)
Visualizing (NaN)
We can use the missingno library to visualize missing data.
Describe categorical variables
mature: ['younger mom' 'mature mom']
premie: ['full term' 'premie']
lowbirthweight: ['not low' 'low']
sex: ['male' 'female']
habit: ['nonsmoker' 'smoker' nan]
marital: ['married' 'not married']
whitemom: ['white' 'not white']Normality check
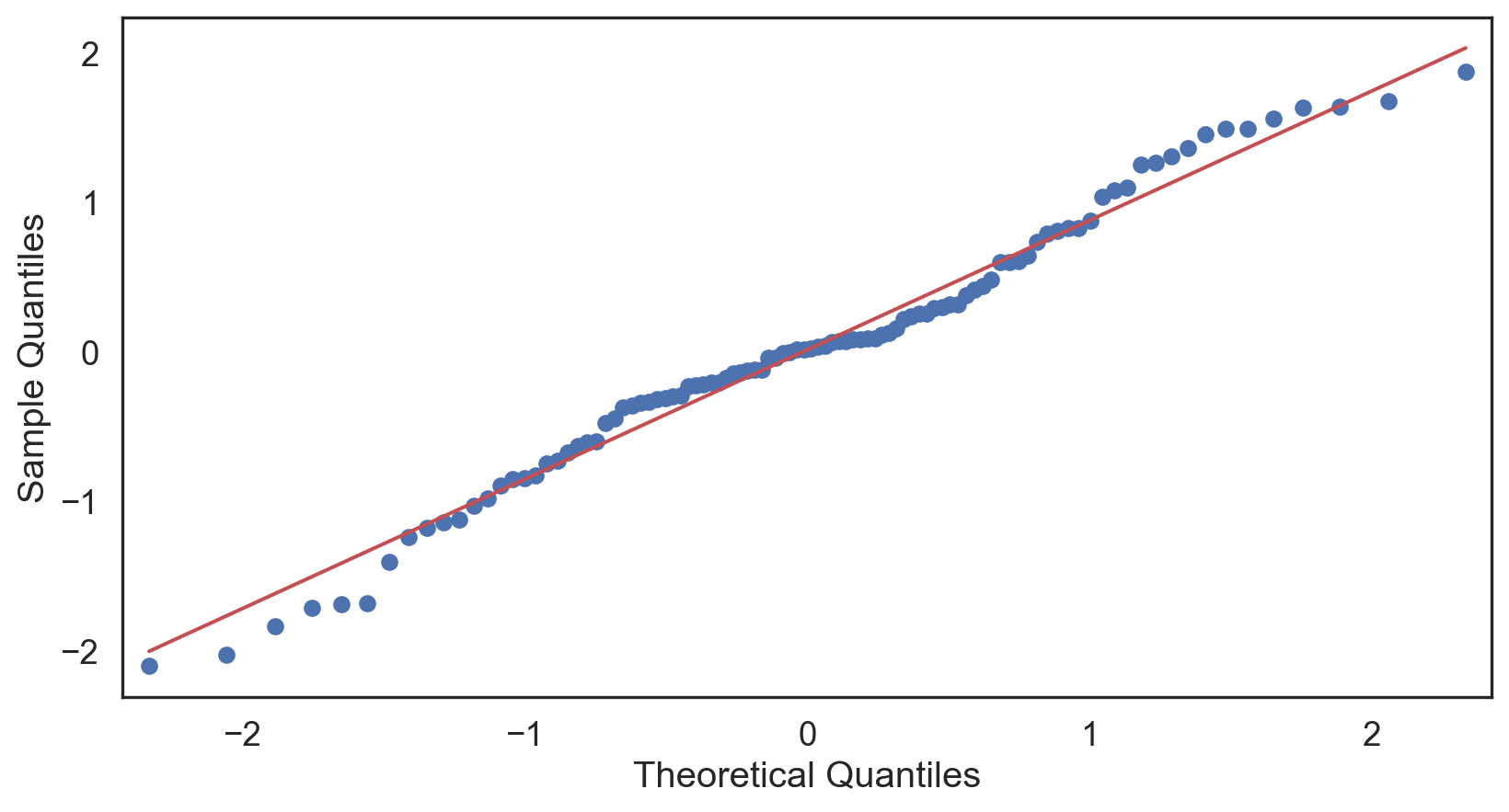
Checking if the data follows a normal distribution is a common step in EDA.
Normality check
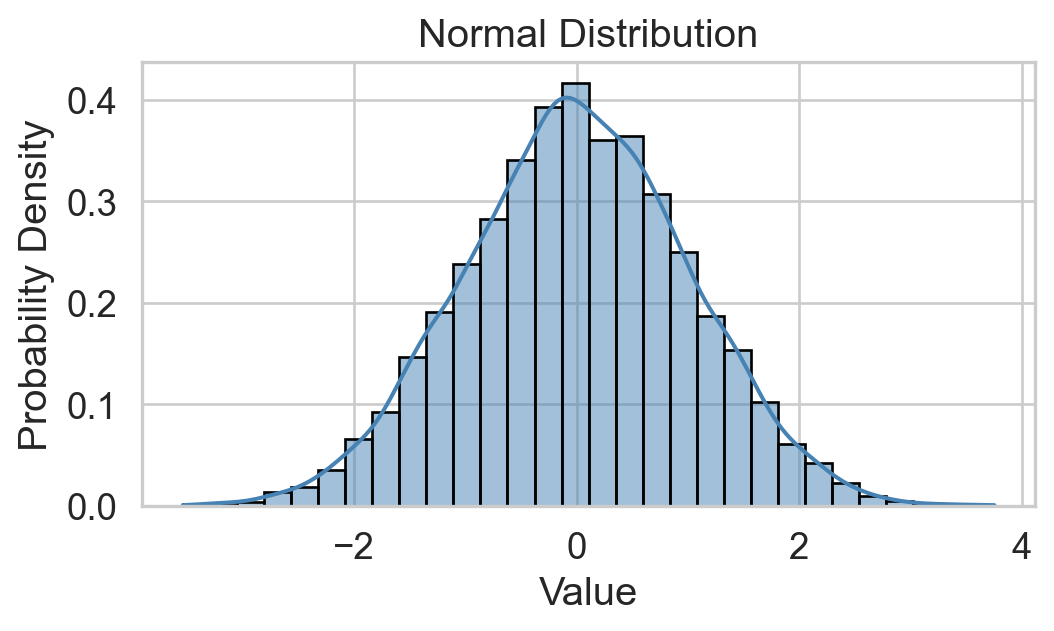
Histogram: bell-shaped curve
Skewness: Close to 0 for symmetry; Kurtosis: Close to 3 for normal “tailedness.”
Sample Size: Larger samples are less sensitive to non-normality.
Empirical Rule: 68-95-99.7% rule (1, 2, and 3 st dev. of the mean).
Skewness
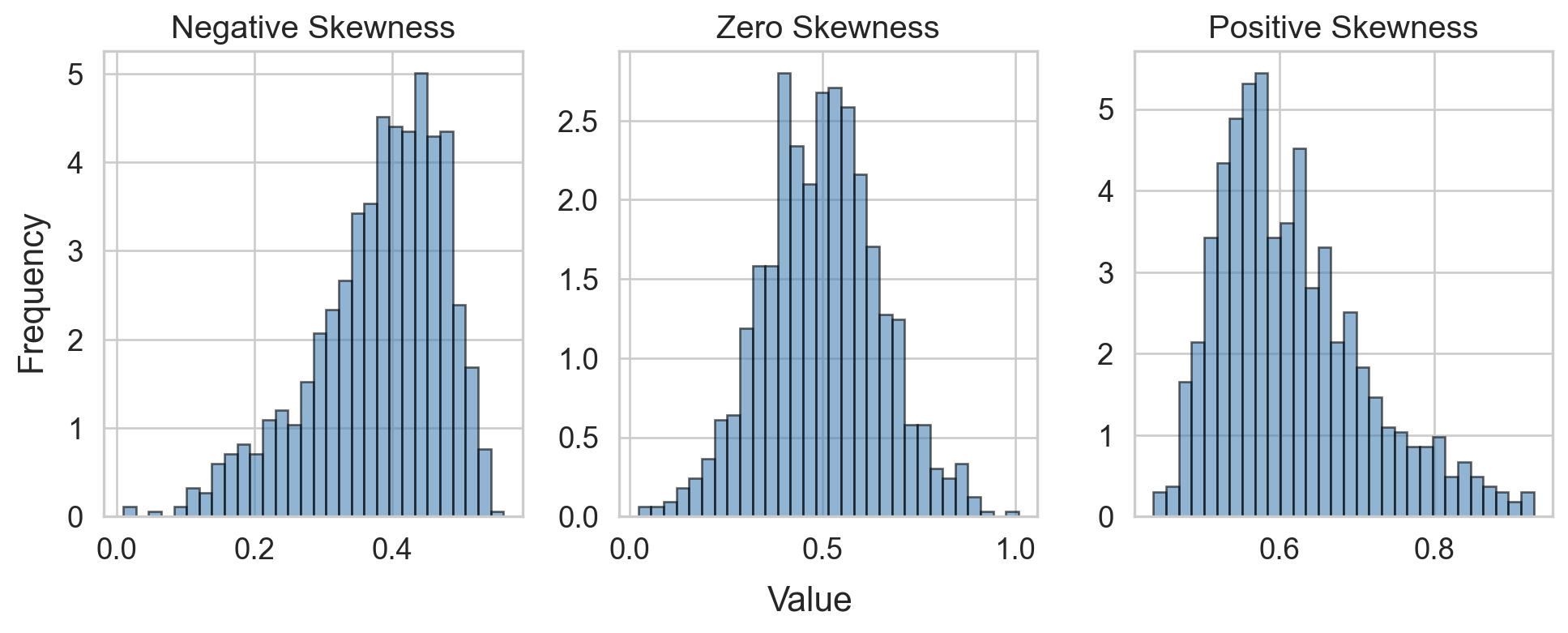
- Several definitions
- Sensitive to outliers
- Designed for one peak (unimodal)
Kurtosis
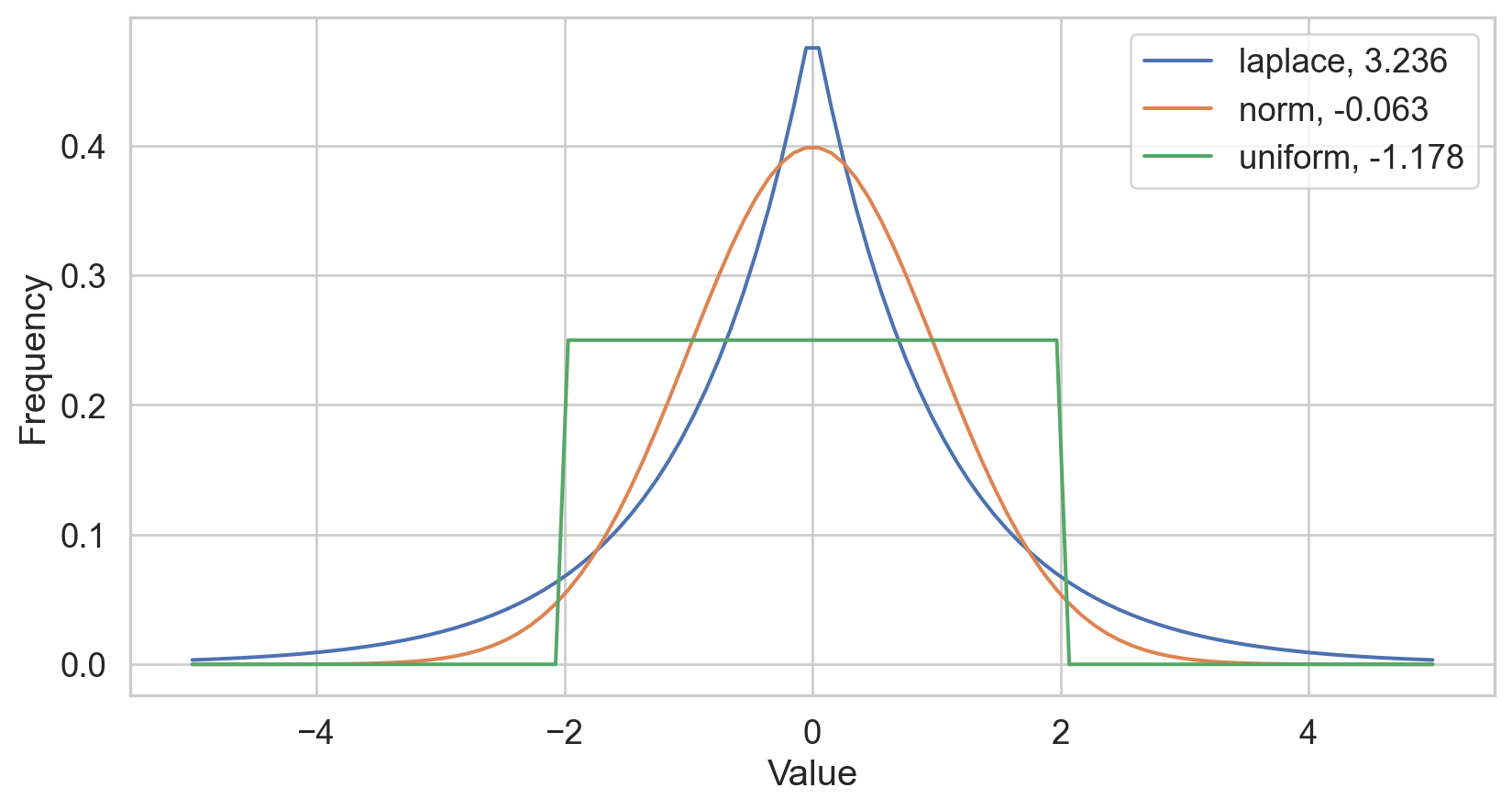
- Sensitive to outliers
- Designed for one peak (unimodal)
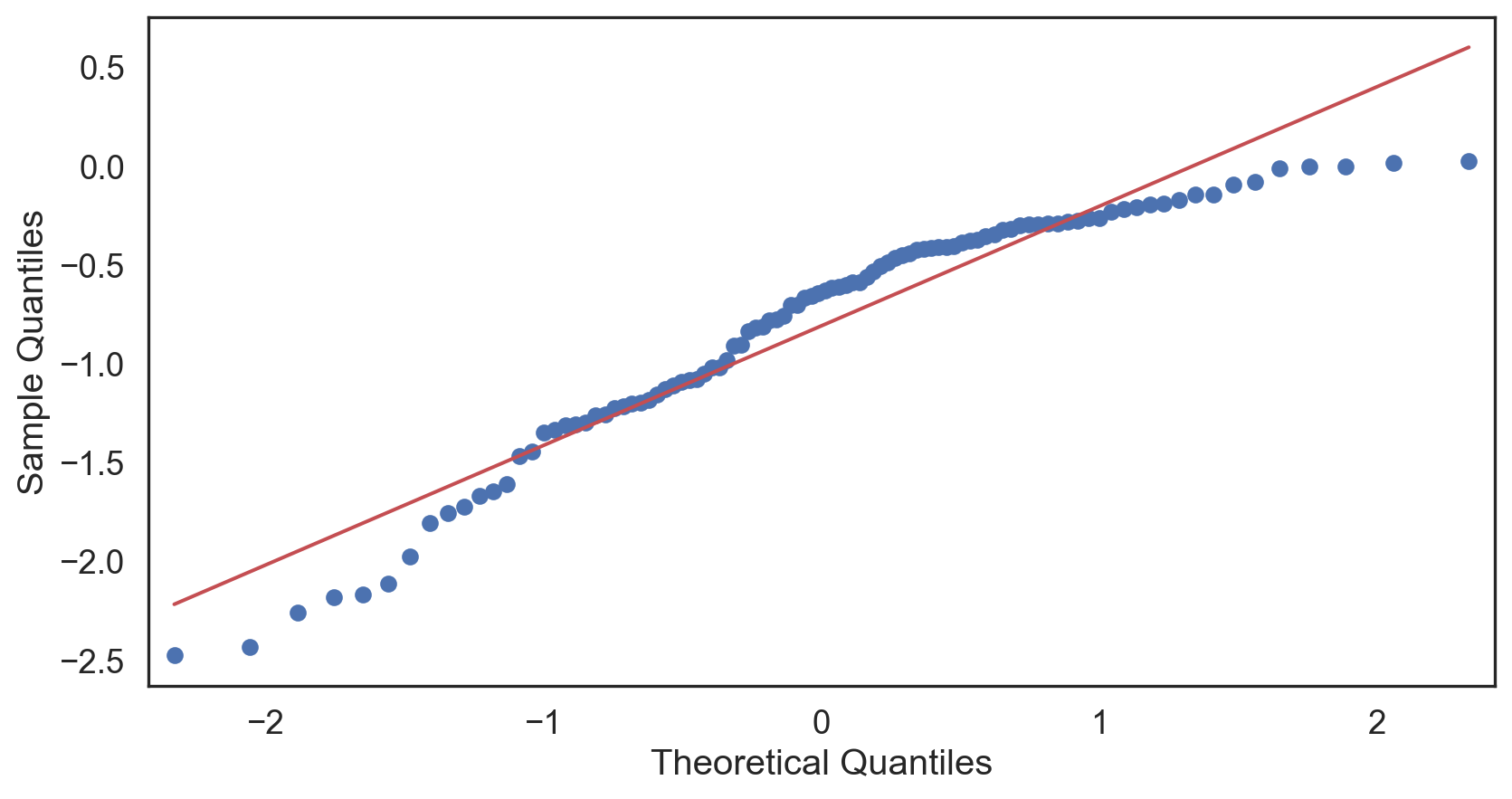
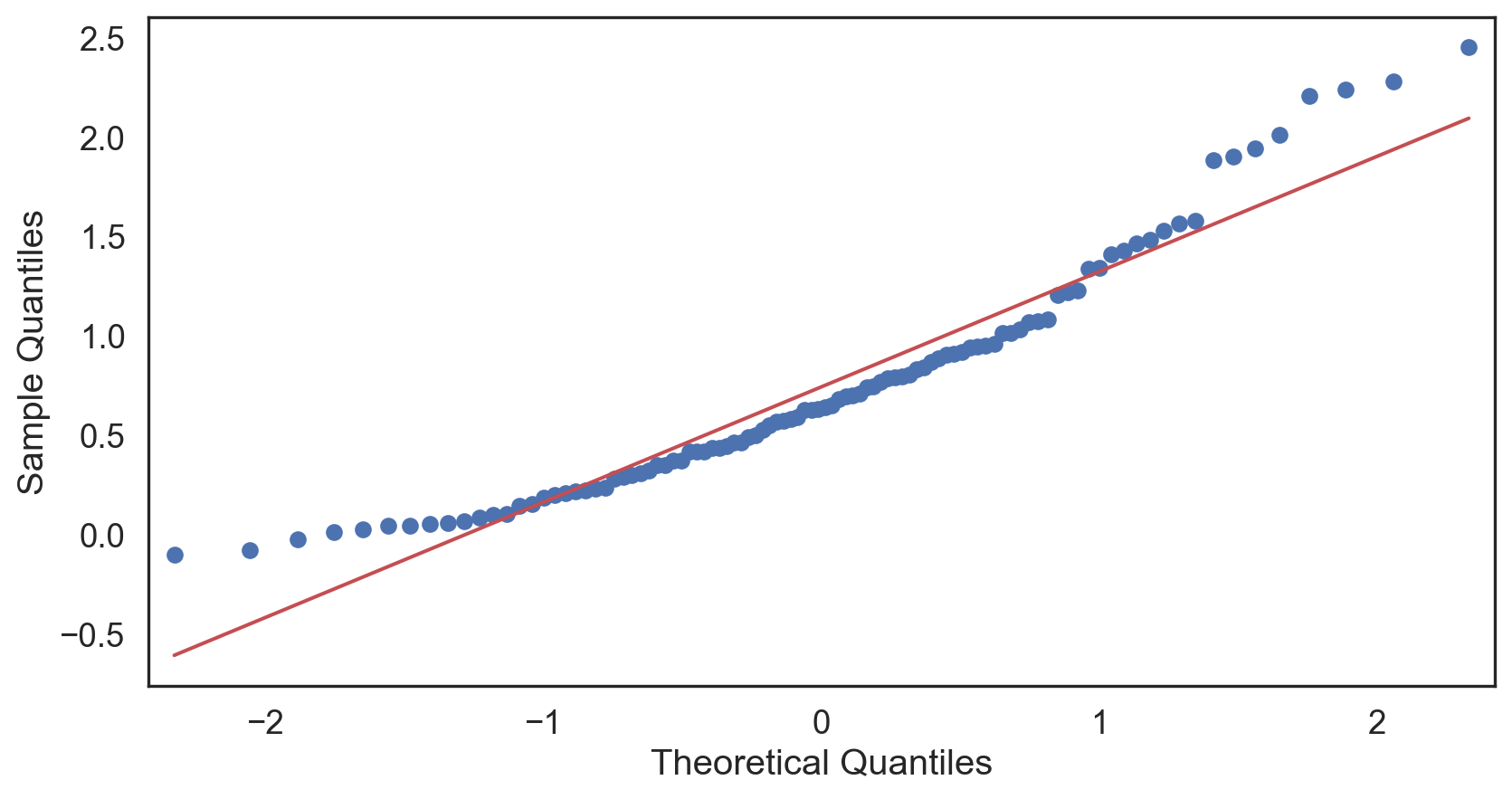
Q-Q plot
Testing normality: data shape
Code
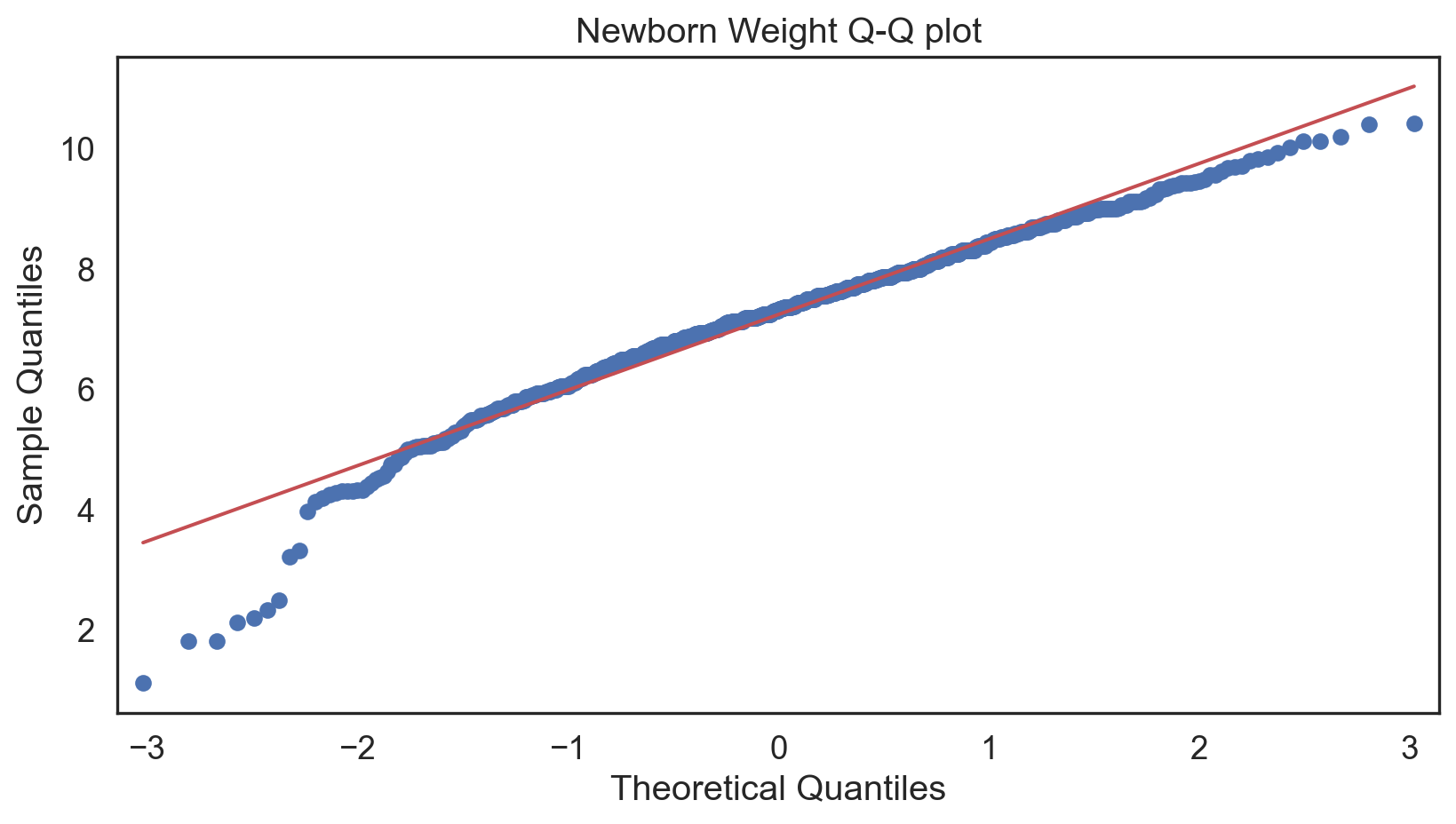
Negative-skew (left-tailed)
Conclusions
Always inspect your data first.
Visualize relationships and distributions.
Identify and handle outliers and missing values.
Check for normality and understand the distribution of your data.